Selasa, 31 Maret 2009
Earth's Dwindling Arctic Sea Ice
You may live to see polar bears become nearly extinct in the wild. On the bright side (if there is one), marine shipping could occur between the Atlantic and Pacific Oceans through the Arctic Ocean during the summer months.
Kamis, 26 Maret 2009
The PSA Test for Prostate Cancer
When the PSA test was first introduced in 1987, scientists thought it might lead to as much as a 50% reduction in prostate cancer deaths. But now it seems that it may not save lives after all. In a study of 77,000 U.S. men, the 10-year death rate from prostate cancer was the same in a group who had an annual PSA test for six years plus a digital rectal examination for four years, compared to a control group who were never tested over the same time period. The study may need to be continued out for another decade or so to determine whether the PSA test has any usefulness over the longer term, however.
How can a test that accurately detects prostate tumors not save lives? Apparently the answer is that most prostate cancers are so slow-growing that older men are likely to die of something else first, even if they do have a diagnosis of prostate cancer. For older men, skipping the PSA test altogether may someday be safe option.
By the way, a digital rectal examination is not some sort of digital readout. It's a physician’s gloved digit, or finger.
Jumat, 20 Maret 2009
If Not Bird Flu, Then.....What?
Knowing this, how might we prevent the next pandemic, or at least have some warning that it was coming? One intriguing possibility would be to keep a close eye on diseases that develop in humans who are in close contact with wild animals. To learn more, read “Preventing the Next Pandemic”, by Nathan Wolfe (Scientific American, April, 2009, pp. 76-81), and then check out the website of the Global Viral Forecasting Initiative. In fact you can download a .pdf file of the Scientific American article directly from the the GVFI website.
Kamis, 19 Maret 2009
Courses, schedule and research opportunities
1/ From Kathy: If you have an advisee looking for a challenging, literature-based, grad seminar style course in S09, I am offering one on the topic of "cell death" and am happy to have ccs students enroll in it through MCDB 194X (2 units). They just need to contact me for an add code. It meets Mondays at 10 AM. I posted a description on the ccs bulletin board and Les sent out an email advertising the course as well.
2/ A general heads up (also from Kathy):
MCDB is working on revising curricula and one of the main goals is to spread out the upper division course offerings across all quarters a bit more (right now, many are bunched in Winter qtr). The most immediate change that might affect our CCS Bio students is that MCDB 112 (Dev Biol) will be offered in FALL 09 (instead of W10), so be aware. The lab (MCDB 112L) will remain a Winter 2010 offering for the moment. Some of the micro courses (such as MCDB 139, medical microbiology) are likely to redistribute as well, and I'll keep everyone posted as I become aware of the changes. For students who are likely to want to take a number of MCDB upper div courses, my advice is to have them get at least 1 qtr of genetics under their belts asap as it seems most faculty really want to see this as a prep for many upper division courses.
3/ A grad student was asking me about whether there were any CCS students interested in a plant based research project. If you are let me know and I'll put you in touch. They were interested in collaborating to put in an application for a Worster award. These awards support the development of graduate and undergraduate research through a mentoring program that pairs an undergraduate with a graduate student mentor during the summer. Stipends this year will be approximately $6000 for each team ($3000 for the graduate and $3000 for the undergraduate). Applications for this award are due by Friday, April 3, 2009.
4/ Sunmer research experience for undergraduates based in Dublin.
Collections-based Biology in Dubin (CoBiD) provides an exciting research environment, with experience both in high quality laboratories in the science departments in University College Dublin, and important international research centre in the National Museum of Ireland (Natural History) collections in Dublin city centre. Diverse research projects are offered, with topics ranging from systematic biology— including traditional and molecular techniques— to ecology and population genetics. Students will work side-by-side with curators and senior scientists and will be involved in all aspects of collections-based research including collection and curation of specimens, participation in field expeditions, and dissemination of scientific results through oral presentation and publication.
The deadline is in two weeks. See website for more info.
Senin, 16 Maret 2009
Posting Drought! Lack of Internet!
Minggu, 15 Maret 2009
Obama Lifts Stem Cell Ban
President Obama’s executive order will permit federally funded researchers to use the hundreds of stem cell lines in existence today, as well as new stem cell lines created by private funding in the future. But there is still a prohibition in place, called the Dickey-Wicker Amendment of 1996, which prohibits federal funding for research “in which human embryos are created, destroyed, discarded, or knowingly subjected to risk of injury or death”. So while federally funded researchers will be able to use stem cell lines created by private funds (because they themselves did not destroy any embryos), they will still be prohibited from creating their own new cell lines from human embryos.
How long can the Dickey-Wicker Amendment can stand up in this new climate of stem cell research permissiveness? It's anybody’s guess. The debate continues…
Sabtu, 14 Maret 2009
Hiatus
Kamis, 12 Maret 2009
SB 4.0 Videos have ARRIVED!!!!!!! (Synthetic Biology 4.0 conference Videos)

It was worth the wait there are over a hundred videos!
Lauren Ha of the BBF and her colleagues from HKUST have uploaded ~110
videos from the SB4.0 conference, held in Hong Kong last October.
The videos are free online via:
http://www.youtube.com/user/BioBricksFoundation
The videos cover many different aspects of the conference, from formal
lectures to informal conversations.
About SB 4.0
The mission of Synthetic Biology 4.0 is to bring together researchers who are working to:
- design and build biological parts, devices and integrated biological systems
- develop technologies that enable such work
- place this scientific and engineering research within its current and future social context
The conference was a coordinated effort between HKUST, Hong Kong University, and Chinese University. Hong Kong provided an ideal location to explore the commercialization of Synthetic Biology in Asia as well as the launching of regional research and educational programs. Further, the meeting facilitated connections between researches and leaders in government, industry, and civic organizations.

Stupid octopus tricks
Well, since I'm not the only one feeling the cephalopod love, here's one more for you. As was noted, octopuses are notorious for escaping from tanks since they are agile, clever and have no rigid skeleton - allowing them to squeeze through practically any gap that they can squeeze their brain through.
This is actually more than just a stupid pet trick since it really helps to remind you that, in water at least, a hydrostatic skeleton, like that of an octopus, can be very impressive and allow sophisticated, rapid and powerful movement.
Rabu, 11 Maret 2009
Reshma Shetty and Natalie Kuldell discuss do-it-yourself biology
Natalie Kuldell
Reshma Shetty
January 14, 2009
Running Time: 1:10:47
Quoted from: http://mitworld.mit.edu/video/646
About the Lecture
Inspired by the vast potential of bioengineering, ordinary people are seeking their inner Frankenstein -- doctor, not monster. Two speakers who know their way around Petri dish and beaker discuss the possibilities and pitfalls of do-it-yourself biology with an MIT Museum crowd.
Showing ads from a 1980 Omni magazine, Natalie Kuldell reflects on the vast changes in computer engineering in the past few decades – from 20-lb PCs to laptops and handhelds. In contrast, she laments, genetic engineering today still resembles in large part its 1980 antecedents -- inserting bits of DNA into organisms like E. coli. She avers that computer engineering made such leaps because its technology was widely available to amateurs, who helped drive many advances. Biotech hasn’t moved as fast, and won’t, believes a nascent do-it-yourself (DIY) community, until basic components of biology become accessible to a larger population.
Synthetic biology aims to make new biological forms easier to engineer. Kuldell complains that “much of my time is spent doing things to do the experiments I need to do. It would be terrific not to have to build things in advance.” But building biological components and streamlining processes is difficult in biology, because biosystems are complex, and unpredictable. Can amateurs working with “Tupperware, thermometers and genetic engineering in the kitchen” discover “something remarkable doing their biology at home?”
Reshma Shetty thinks engineered organisms can do more than sense toxic metals in the environment or determine whether seawater is contaminated. She can “imagine a DIY bioengineer…doing something more fantastical, ambitious…. What about growing your own house?” Shetty describes a home experiment that can make bacteria smell like bananas. This is a small feat, but to achieve something significant, a real contribution to science, Shetty says DIY biologists need bio-engineered friendly organisms that will serve as common models, safe, easy to grow “and fun to use.” Candidates include moss, an easy to grow bacterium called Acinetobacter, and the salt-loving Halobacterium. By giving people the right tools, “they can build something fun and creative others can appreciate.”
Natalie Kuldell

Instructor of Biological Engineering, MIT
Natalie Kuldell did her doctoral and post-doctoral work at Harvard Medical School. She develops discovery-based curricula drawn from the current literature to engage undergraduate students in structured, reasonably authentic laboratory experiences. She has also written educational materials to improve scientific communication as it occurs across disciplinary boundaries and as it's taught in undergraduate subjects. Her research examines gene expression in eukaryotic cells, focusing most recently on synthetic biology and redesign of the yeast mitochondria. She serves as Associate Education Director for SynBERC, an NSF-funded research center for Synthetic Biology, and Councilor at Large for the Institute of Biological Engineering.
Reshma Shetty PhD '08

Founding Member, Ginkgo Bioworks
Reshma Shetty earned her MIT Ph.D. in Biological Engineering, where she engineered bacteria to smell like mint and banana. She has been active in the field for several years and co-organized SB1.0, the first international conference in synthetic biology in 2004. She spearheaded the use of OpenWetWare, a wiki for life science researchers, as an educational tool when she helped teach an MIT undergraduate laboratory course in synthetic biology in 2006. The course demonstrated how wiki’s can support university education and has served as a model for courses from institutions across the country. She also engineered bacteria to smell like mint and banana’s. Now she and four other MITers have founded a new synthetic biology startup called Ginkgo BioWorks.
Octopuses in disguise
Once you start posting cephalopod videos it's hard to stop. This one is a nice overlap between diversity and next quarter's physiology though.
Getting a paper published in science as a grad student. Excellent.
Giving it a cool title: Underwater Bipedal Locomotion by Octopuses in Disguise. Even better.
Capturing the whole thing on video. Priceless.
A Way to Cure HIV Infection?
A new approach is anti-latent therapy – therapy designed to prevent dormant viruses from staying dormant and hidden. The idea is to force any remaining dormant viruses to become active again so that they can be targeted and killed by suppressive therapy. A combination of anti-latent therapy and suppressive therapy just might wipe out an HIV infection completely. At least, that’s the idea.
Selasa, 10 Maret 2009
Futures in Biotech 8: Drew Endy on Synthetic Biology (must listen! webcast from 2006)


If you haven't heard this yet, it is an absolute must listen. It is from 2006 but it's a wonderful presentation from Drew. One of my favorites. The ability of Drew to dissect an argument down to it's core concepts makes my inner philosopher jump for joy. Sadly, I can't embed so you'll have to click here: http://www.twit.tv/fib8
J. Chris Anderson, asst. professor of bioengineering at UC Berkeley, leads a discussion on synthetic biology. (Feb 16th 2009 video)
J. Chris Anderson, asst. professor of bioengineering at UC Berkeley, leads a discussion on synthetic biology. The discussion starts with a detailed overview of synthetic biology (and its distinction from genetic engineering). Chris then dives into the meat of his work, engineering E. Coli strains to target cancer cells.
Additional Resources
Chris’ Lab Page
Cthulhu hates Chordates
 Having just enthused about Cephalopods I was delighted to come across this picture in the news today. I'm a big fan of obscure jokes and this placard, at a counter demonstration to the hateful Fred Phelps, who was at the University of Chicago yesterday, is excellent. Fred Phelps and his church, the Westboro Baptist Church are (in)famous for their protests which usually involve a lot of placards along the lines of 'God hates gays', 'God hates the world' or 'God hates you'.
Having just enthused about Cephalopods I was delighted to come across this picture in the news today. I'm a big fan of obscure jokes and this placard, at a counter demonstration to the hateful Fred Phelps, who was at the University of Chicago yesterday, is excellent. Fred Phelps and his church, the Westboro Baptist Church are (in)famous for their protests which usually involve a lot of placards along the lines of 'God hates gays', 'God hates the world' or 'God hates you'.This parody is great since it requires you to know something about HP Lovecraft to know that Cthulhu is very Cephalopod like AND to know that Cephalopods are most decidedly not Chordates. In addition, the Lovecraft Cthulu WAS evil and probably does hate Chordates. Kudos.
MTA. A comment from the signmaker: I decided that if the Mighty Tentacled One had to hate a chosen group, it would be one outside of it's representative clade, and deuterostomes wouldn't fit on a sign.
Cephalopod star
Of course as biologists we want to look a little deeper. The cool bit is that it gets even more amazing as you think about it more closely. As PZ Myers pointed out on his blog, the octopus needs to do four things to achieve this trick:
- It needs good visual system. In order to match the background you need to be able to see it. To match it well, you need to see it well.
- To pull off the fast change you need a fast connection from the brain to the color changing organ.
- Speaking of which, yes, the octopus needs organs that can change color. Cephalopods have tiny, discrete sacs of pigment scattered all over their body, each one ringed with muscles that can close the sac to conceal the pigment, or expand the sac to expose the pigment.
- Finally, the octopus needs a set of rules, an algorithm, so it can translate what is sees with its eyes into a visual pattern that hides the animal.
Today is Cynthia Kenyon day! Her work has led to the discovery that the aging process is regulated by our genes! (video, webcast, and a summary)


In 1993, Dr. Kenyon's discovery that a single-gene mutation could double the lifespan of C. elegans sparked an intensive study of the molecular biology of aging. Dr. Kenyon's findings have led to the discovery that an evolutionarily-conserved hormone signaling system influences aging in other organisms, including mammals. Kenyon has received many honors, including the King Faisal Prize for Medicine, the American Association of Medical Colleges Award for Distinguished Research, the Ilse & Helmut Wachter Award for Exceptional Scientific Achievement, and La Fondation IPSEN Prize, for her findings. She is a member of the U.S. National Academy of Sciences and the American Academy of Arts and Sciences. She is now the director of the Hillblom Center for the Biology of Aging at UCSF.
Cynthia Kenyon gives us an overview of her labs work:
"Aging and death are always with us. The sense of loss that comes with aging and death imbues the sonnets of Shakespeare, the stories of Oscar Wilde and the art of Cranach and others with great meaning and beauty. The idea of a fountain of youth is enchanting, but it has always been the stuff of fairy tales, not science. Scientists, too, think about aging, and they have been studying the aging process for a long time. But, like non-scientists, most of them have assumed that while it might be possible to live longer with a healthier lifestyle, nothing much could be done to fundamentally change the rate of aging.
Some of the most important discoveries in science have come not from studying people themselves, but from studying simpler creatures: bacteria, yeast, roundworms, fruit flies and mice. Although these animals look very different from one another and from people, they share universal mechanisms of life at the molecular level.
My lab has been studying a small microscopic roundworm called C. elegans for some time, and these animals are perfect for studies of aging because they get old and die in just a little over two weeks. What is more, it’s easy to look for genes that control virtually any process, simply by changing them (making mutations) and looking at the consequences.
It seemed to me that there was a good chance that the aging process, like so much else in biology, was not just a random and haphazard process but instead was subject to regulation by the genes. After all, rats live three years and squirrels can live for twenty-five, and these animals are different because of their genes. Also, most biological processes are subject to tight control by the genes. If so, then by finding genes that control aging, and then changing the activities of the proteins they encode, one day we might be able to stay young much longer than we do now.
When we began our studies of aging, in the early 1990s, one C. elegans gene that affected lifespan had been described, though it was poorly understood. When this gene was altered by a mutation, the animals lived 30-50% longer than normal. We looked for gene changes (mutations) that extended the lifespan of the roundworms, and we found that mutations in a gene called daf-2 doubled lifespan. These mutant worms still looked and acted young when they should be old. Seeing them was like talking to someone that looks 40 and learning that they were really 80. This was a stunning finding because no one thought it was possible. We also discovered another important gene, called daf-16, that was needed for this long lifespan. daf-16 was a gene that could keep an animal young.
We now know that these genes, daf-2 and daf-16, allow the tissues to respond to hormones that affect lifespan. We showed that daf-2 and daf-16 ultimately affect lifespan by influencing the activities of a wide variety of subordinate genes that influence the level of the body’s antioxidants, the power of its immune system, its ability to repair its proteins, and many other beneficial processes. We have found that the activity of the youthfulness gene daf-16 is influenced by signals from the environment and also by signals from within - from its reproductive system. This knowledge has now allowed us to extend the lifespan of active, youthful worms by six fold.
Others have now extended these findings to show that daf-2-like genes control the lifespan of fruit flies, mice and possibly (from studies that will be published soon) also humans. When these genes are changed, aging is slowed and lifespan is extended.
Especially wonderful is the fact that these long-lived animals are resistant to a variety of age related diseases, including (in various animals) cancer, heart failure, and protein-aggregation disease. Thus these mutants not only look young, they are young, in the sense that they are not susceptible to age-related disease until later. (Many people assume that if you could delay aging, you would just die of Alzheimer’s disease. We don’t know for sure, but this may not be true if ‘being elderly’ is what makes one susceptible to Alzheimer’s disease.) This link between aging and age-related disease suggests an entirely new way to combat many diseases all at once; namely, by going after their greatest risk factor: aging itself. This is an extremely exciting and important concept that could revolutionize medicine, human health and longevity, and it has just now begun to be studied in earnest, still in only a handful of labs.
Because it is very easy to look for genes affecting lifespan in C. elegans, we are continuing to do that in our lab. In fact, you can think of C. elegans as a ‘fountain of youthfulness genes’. We have identified about fifty genes so far that affect lifespan, and others have found this type of gene as well. More importantly, we are now using all the powerful molecular techniques available for studying this little animal to figure out just what these genes do to affect lifespan, so that we can apply that knowledge in a rational way. Whether these genes have universal effects on lifespan can now be tested in higher animals, where it is harder to discover lifespan genes starting from scratch. With all this new information, pharmaceutical and biotech companies can now make drugs that influence the activities of the proteins encoded by these genes, in hopes of combating age-related disease, and possibly aging itself, in humans. We don’t know yet, but to me it seems possible that a fountain of youth, made of molecules and not simply dreams, will someday be a reality."
To hear more from Cynthia Kenyon you can listen to her interview with Marc Pelletier on the webcast, Futures in Biotech 36: Avoiding Death, Not Taxes with Dr. Cynthia Kenyon Published on Nov 24, 2008
Host: Marc Pelletier Guest: Dr. Cynthia Kenyon; Professor, Department of Biochemistry and Biophysics, University of California San Francisco, Director of the Larry L. Hillblom Center for the Biology of Aging. We are back into a world leading lab to discuss the genetics of aging. Can it be controlled? You bet, and the implications are enormous. When these findings translate to the clinic, it will truly be a game changer for humanity.
You can also hear Cynthia Kenyon's American Society for Cell Biology iBio Seminar on Aging here:

And if you haven't seen it already, make sure to check out her discussion at the 2007 Aspen Health Forum: "Science vs the Biological Clock"
William Colby, Cynthia Kenyon and Stephanie Lederman all discuss the process of aging:
http://diybio4beginners.blogspot.com/2009/02/science-versus-biological-clock-video.html
Senin, 09 Maret 2009
INTRO TO CADHERINS AND INTEGRINS - THE GLUE THAT STOP US FROM BEING GOO! (Videos + Seed Magazine Article)
Excellent Videos about Cadherins (I was not able to embed the videos so click on the links below):
Cadherins: Structure and Function Part I ( introduction)
Cadherins: Structure and Function Part II ( Cadherin Molecular Structure and Function )
Cadherins: Structure and Function Part III ( Adherens Junctions and Tissue Morphogenesis )
Cadherins: Structure and Function Part IV ( Cadherins in the Neural Network )
Cadherins: Structure and Function Part V ( Conclusion )

EXCERPT from Seed Magazine article: The Mason's Apprentice
http://www.seedmagazine.com/news/2008/10/the_masons_apprentice_1.php
"Multicellularity requires complex cell adhesion and signaling abilities — development and differentiation cannot occur without them. A multicellular organism is made up of cells that stick to one another with varying degrees of strength, which is mediated by an external coat of proteins and sugars that makes cells sticky in specific ways. In addition, cells secrete proteins and sugars that form a kind of fibrous goo called the extracellular matrix, to which they can also stick. When cell proteins bind to other cells or the extracellular matrix, the proteins trigger biochemical changes — the signaling part of the process — that can cause changes in cell metabolism, gene activity, cell shape, and physiology. These capabilities are fundamental to building a multicellular organism.
So where did they come from?
One must be careful when investigating this question not to make an easy but erroneous assumption: that cell adhesion and cell-to-cell signaling are a consequence of multicellularity. They are not. In fact, it turns out that single-celled organisms have a diverse array of mechanisms for interacting with one another, and multicellular life's fancy cell-communication tools are recent appropriations of mechanisms refined by evolution over billions of years, well before the first tiny worm congealed in the late pre-Cambrian.
"Simple" one-celled organisms like bacteria (which aren't simple, except in terms of number of cells) are sensitive to their environment, including the presence of other bacteria, and transduce chemical signals around them into changes in gene activity. The central principles of cell signaling are all in place in E. coli, and we can see the general idea clearly expressed in the rest of the prokaryotes. But another group of single-celled organisms, a group of eukaryotes — are of particular interest to multicellular animals like ourselves because they are the protists most closely related to us. These organisms are pf great interest to evolutionary biologists because they demonstrate that our toolbox of cell-adhesion and signaling proteins are of utility to organisms that don't have tissues and a higher level of organization. These fascinating creatures are the choanoflagellates.
Two particularly significant classes of proteins that animals use for adhesion and signaling are shared between animals and choanoflagellates. One is a group of proteins called cadherins. These are important cell-adhesion molecules that are regulated by calcium in the environment. Before being found in choanoflagellates, cadherins were thought to be unique to animals — plants and fungi do not have them. Another is a group of proteins called integrins that help cells stick to the extracellular matrix. Among other things, these molecules adhere to the collagen in connective tissues; they are essential for holding us together in a coherent form, versus a pile of gooey jelly."
Wiki:
Cadherins are a class of type-1 transmembrane proteins. They play important roles in cell adhesion, ensuring that cells within tissues are bound together. They are dependent on calcium (Ca2+) ions to function, hence their name. The cadherin superfamily includes cadherins, protocadherins, desmogleins, and desmocollins, and more. In structure, they share cadherin repeats, which are the extracellular Ca2+-binding domains.
There are multiple classes of cadherin molecule, each designated with a prefix (generally noting the type of tissue with which it is associated). It has been observed that cells containing a specific cadherin subtype tend to cluster together to the exclusion of other types, both in cell culture and during development. For example, cells containing N-cadherin tend to cluster with other N-cadherin expressing cells. However, it has been noted that the mixing speed in the cell culture experiments can have an effect on the extent of homotypic specificity.[1] In addition, several groups have observed heterotypic binding affinity (i.e., binding of different types of cadherin together) in various assays.[2][3] One current model proposes that cells distinguish cadherin subtypes based on kinetic specificity rather than thermodynamic specificity, as different types of cadherin homotypic bonds have different lifetimes.[4]
Different members of the cadherin family are found in different locations. E-cadherins are found in epithelial tissue; N-cadherins are found in neurons; and P-cadherins are found in the placenta. T-cadherins have no cytoplasmic domains and must be tethered to the plasma membrane.E-cadherin (epithelial) is the most well-studied member of the family. It consists of 5 cadherin repeats (EC1 ~ EC5) in the extracellular domain, one transmembrane domain, and an intracellular domain that binds p120-catenin and beta-catenin. The intracellular domain contains a highly-phosphorylated region vital to beta-catenin binding and therefore to E-cadherin function. Beta-catenin can also bind to alpha-catenin. Alpha-catenin participates in regulation of actin-containing cytoskeletal filaments. In epithelial cells, E-cadherin-containing cell-to-cell junctions are often adjacent to actin-containing filaments of the cytoskeleton.
E-cadherin is first expressed in the 2-cell stage of mammalian development, and becomes phosphorylated by the 8-cell stage, where it causes compaction. In adult tissues, E-cadherin is expressed in epithelial tissues, where it is constantly regenerated with a 5-hour half-life on the cell surface.
Loss of E-cadherin function or expression has been implicated in cancer progression and metastasis. E-cadherin downregulation decreases the strength of cellular adhesion within a tissue, resulting in an increase in cellular motility. This in turn may allow cancer cells to cross the basement membrane and invade surrounding tissues.
Another is a group of proteins called integrins that help cells stick to the extracellular matrix.Heat Stress Causes Bacterial Forsight - SEED MAGAZINE Article
http://www.seedmagazine.com/news/2008/10/bacterial_foresight.php
Can bacteria anticipate changes in their environment?
The homeostatic framework has long dominated the study of bacteria and microbiology, asserting that bacteria change their behavior based on the information they receive from their local environment. Researchers know, for example, that when E. coli bacteria enter the gut — an environment lacking oxygen — they switch to a form of anaerobic respiration in order to survive.
But there is a fundamental problem for any organism that behaves only by reacting to its environment after the fact: The behavior is not very efficient. If bacteria had the ability to use environmental cues to plan for future changes, the transition would be far smoother, and their survival more assured.
A group of microbiologists studying E. coli recently noted that before entering the deoxygenated gut, the bacteria enter the mouth and experience a rise in temperature. When the researchers exposed the bacteria to a similar increase in temperature, as if in anticipation of entering the gut, they found that E. coli turned to anaerobic respiration even without oxygen deprivation.
See also:
Predictive behavior within microbial genetic networks
Science 6 June 2008
Seminars
Monday March 9th, 4-5pm, MSRB Auditorium
"Variation in Resilience and Species Interactions Among Southern Californian Kelp Forest Ecosystems"
Anne Salomon
Tuesday, March 10, from 8 to 9 p.m in Broida Hall, Room 1610.
Public Lecture and Demonstrations
'Journey to the Heart of the Electromagnetic Spectrum'
Would you like to know how doctors might one day be able to see through bandages? To learn about this and many other technological breakthroughs, you are invited to attend "A Journey to the Heart of the Electromagnetic Spectrum." The event is designed for anyone who is curious, regardless of technical background, and will be presented by Mark Sherwin, Professor of Physics and Director of the newly formed Institute for Terahertz Science and Technology.
Tuesday, March 10, ESB 1001, 11:00 AM
Mellichamp Chair in Systems Biology Search Seminar
"Nature, nurture or just dumb luck: gene expression variability and cell fate"
Arjun Raj, Department of Physics, Massachusetts Institute of Technology, Cambridge, MA
Thursday March 12, MCDB Seminar , 3:30pm LSB Auditorium (1001)
“Dying Young as Late as Possible: Regeneration, Planarians and Stem Cells”
Alejandro Sánchez Alvarado, Ph.D.
Howard Hughes Medical Institute, Dept. of Neurobiology & Anatomy
University of Utah School of Medicine
Convergence: Synthetic Biology Panel (2008 video)
Convergence: Synthetic Biology Panel (Part 1 of 2) from Jeriaska on Vimeo.
(Part 2 of 2)
Convergence: Synthetic Biology Panel (Part 2 of 2) from Jeriaska on Vimeo.
About the speakers:
Chris Anderson is a bioengineering researcher and educator. He received his Ph.D. in 2003 from the Scripps Research Institute for expanding the genetic code through genetic engineering. Currently he is a professor in the Department of Bioengineering at UC Berkeley. His research focuses on foundational technologies and applications of synthetic biology, a ground-up approach to genetic engineering with diverse applications in healthcare, environmental remediation, bioenergy, chemicals and materials production. Chris is best known for his ongoing work on developing therapeutic bacteria for the treatment of cancer for which he was recognized with Technology Review's TR35 award in 2007.
Denise Caruso co-founded the nonprofit Hybrid Vigor Institute in 2000 to study and practice collaboration in the service of new solutions for complex social and scientific problems. She recently published Intervention: Confronting the Real Risks of Genetic Engineering and Life on a Biotech Planet, and continues to work on projects both in academia and the private sector to improve the practice of risk analysis for science and technology-related innovations. For the five years prior to founding Hybrid Vigor, Denise wrote the Technology column for the Monday Information Industries section of The New York Times.
Gregory Benford is a physicist, educator, and author. He received his Ph.D. in 1967 from UC San Diego. Benford is a professor of physics at UC Irvine, where he has been a faculty member since 1971. He conducts research in plasma turbulence theory and experiment, and in astrophysics. He has published over a hundred papers in fields of physics from condensed matter, particle physics, plasmas and mathematical physics, and several in biological conservation. He is a Woodrow Wilson Fellow at Cambridge University, and has served as an advisor to the Department of Energy, NASA and the White House Council on Space Policy. In 1995 he received the Lord Foundation Award for contributions to science and the public comprehension of it. He is the author of over 20 novels, including Jupiter Project, Artifact, Against Infinity, and Timescape. He is a two-time winner of the Nebula Award.
Andrew Hessel, MSc, iGEM Program Development, Alberta Ingenuity Fund, is a biologist and author working to promote synthetic biology and open source biology. In his view, synthetic biology allows forward engineering, permitting scientists to write code de novo, and allowing logical, fully understandable evolution of biological outputs ranging from single proteins to synthetic bacteria. Andrew advocates the use of open source principles for creating DNA code. He believes open biology could potentially create a more diversified and sustainable biotechnology industry.
DIY BIO COMMUNITY OVERVIEW IN 4 MINS WITH MAC COWELL
The DIYbio Community - Presented at Ignite Boston 5 (2009) from mac cowell on Vimeo.
Quoted from diybio.org...
"We founded diybio.org, a community for amateur scientists, last year in May, just in time to present at ignite boston 2008. Since then, the community has grown. In this talk, I spend 5 minutes giving a lighting overview of the community and the current hot projects members are working on: new, cheap, diy-hardware, distributed science experiments (think flashmobs for science), a biohacking coworking space, and some molecular biology experiments (including making genetically engineered fluorescent yogurt, a melamine biosensor, and a biological counter)."
Mac's first video at O'Reilly Ignite Boston 2008:
DIYbio in 5 minutes - O'Reilly ignite Boston from mac cowell on Vimeo.
Minggu, 08 Maret 2009
Wolfram Alpha
 Kathy sent me some information about a new tool from Wolfram (the people that make Mathematica). It is called Wolfram Alpha and promises to change the way we use the internet for research. At the moment there is more hype than actual information but Kathy saw a demo and was impressed. Basically it looks like a search engine but instead of just searching for websites with keywords it will take questions in natural language and return answers, provided the data is available, even if no-one has addressed that question before.
Kathy sent me some information about a new tool from Wolfram (the people that make Mathematica). It is called Wolfram Alpha and promises to change the way we use the internet for research. At the moment there is more hype than actual information but Kathy saw a demo and was impressed. Basically it looks like a search engine but instead of just searching for websites with keywords it will take questions in natural language and return answers, provided the data is available, even if no-one has addressed that question before.There is clearly a huge market for such a tool. Currently there is a ton of data out there on the internet - but unless someone has already asked your question you are out of luck in getting a quick easy answer. Say I want to know the relationship between seed size and mature plant size, or how stomatal density varied over the Cenozoic, or how VO2 max varies with animal body size. Unless someone has already asked these questions I would be left to combine data sets on my own. If this product lives up to its promise it will present the data to me - and in graphical form. Cool. I'm sold. I like the last line in the blog posting:
I think it’s going to be pretty exciting. A new paradigm for using computers and the web.
That almost gets us to what people thought computers would be able to do 50 years ago!
Sabtu, 07 Maret 2009
Things Darwin would not say
 In honor of the 200th anniversary of Darwin's birth, New Scientist magazine had a competition to come up with the best 'Thing you would never have heard Charles Darwin say about evolution.' Out of 900 entries the winner was:
In honor of the 200th anniversary of Darwin's birth, New Scientist magazine had a competition to come up with the best 'Thing you would never have heard Charles Darwin say about evolution.' Out of 900 entries the winner was:'Finches eh? Seen one, seen 'em all'
Also in New Scientist this week The Last Word, where readers send in questions and then others submit answers, has:
I have always been fascinated by evolution, and while I can usually see why and how certain characteristics evolved in different species, I'm confused by whales and dolphins. How did their breathing holes evolve, bearing in mind their ancestors were land mammals?
You can see the answers submitted to date here.
AGBT 2009 – Notes (Sadly I could find no other videos)
http://www.fejes.ca/labels/AGBT%202009.html
Next-Generation Informatics - Fresh Video from Feb 2009 AGBT conference
David Dooling recreates the talk he gave in the Bioinformatics session at AGBT 2009.

2009 AGBT Meeting
"The 10th annual Advances in Genome Biology and Technology (AGBT) meeting will be held in Marco Island, Florida, from February 4-7, 2009. The AGBT meeting has become the premier scientific forum for capturing the latest advances in new DNA sequencing technologies and an outstanding venue for presentations on the applications of genomics to diverse areas in biology and biomedicine."Biology Prefixes and Suffixes

Biology Prefixes and Suffixes that start with A or B are below... you can find the rest of the alphabet here: http://biology.about.com/od/prefixesandsuffixes/a/aa020106a.htm
A and B
SUFFIX
-ase = enzyme
Examples: sucrase (sucr-ase) - an enzyme that catalyzes the decomposition of sucrose into glucose and fructose
-ate = having, characterized by, resembling
Examples: nervate (nerv-ate) - leaves characterized by prominent veins
-ary = of or relating to
Examples: urinary (urin-ary) - of or relating to urine and its production or excretion
PREFIX
auto- = self. Examples: autotroph (auto-troph) - organism that is self nourishing or capable of generating its own food
asco- = sac, bag. Examples: ascomycete (asco-mycete) - fungi whose spores are produced in a sac
arth- = joint. Examples: arthritis (arth-itis) - joint inflammation
antho- = flower. Examples: anthophyta (antho-phyta) - plant division composed of flowering plants
ante- = before. Examples: antemortem (ante-mortem) - before death
angio- = vessel. Examples: angiotensin (angio-tensin) - neurotransmitter that causes blood vessels to become narrow
andro- = male. Examples: androgen (andro-gen) - male hormone
ana- = upward, back, again. Examples: anaplasia (ana-plasia) - cell reverting to an immature form
-amyl = starch. Examples: amylase (amyl-ase) - a group of starch enzymes
amphi- = both, on both sides, around. Examples: amphibian (amphi-bian) - animal that can live on both land and water
ambi- = both. Examples: ambidextrous (ambi-dextrous) - capable of using both hands
aer- or aero- = air, oxygen. Examples: aerobic (aer-o-bic) - with oxygen
ad- = toward, near. Examples: adrenal (ad - renal) - toward the kidneys
ab- = away from. Examples: abnormal (ab - normal) - departing from normality
a- = without, negative, not. Examples: asexual ( a- sexual) - without sex
bi- = two. Examples: biennial (bi-ennial ) - plant with two year life span
bio- = life. Examples: biology (bio-logy) - the study of life
brachio- = upper arm, forelimb. Examples: brachium (brachi-um) - arm-like part of an animal
brady- = slow. Examples: bradycardia (brady-cardia) - slow heart beat
bronchi- = windpipe. Examples: bronchioles (bronchi-oles) - small tubes in the lungs
bryo- = moss. Examples: bryophyte (bryo-phyte) - mosses
-blast = bud or germ. Examples: osteoblast (osteo-blast) - a cell from which bone is derived
FOR C-Z click here -> http://biology.about.com/od/prefixesandsuffixes/a/aa020106a.htm
The ecosystem that is your mouth! The Human Oral Microbiome by Floyd Dewhirst, Harvard University
"The human oral cavity is a diverse habitat that contains approximately bacterial 600 predominant species. The oral microbiome is comprised of 44% named species, 12% isolates representing unnamed species, and 44% phylotypes known only from 16S rRNA based cloning studies. Species from 11 phyla have been identified: Firmicutes (211), Bacteroidetes (106), Proteobacteria (99), Actinobacteria (64), Spirochaetes (49), Fusobacteria (29), TM7 (12), Synergistetes (10), Chlamydiae (1), Chloroflexi (1) and SR1(1). Full and survey sequences have been obtained for over 30 oral species, and in the course of the Human Microbiome Project over 300 essentially complete genome sequences should be determined. An Oral Microbiome Project is in progress and data from this project should be available soon. The talk will discuss the diversity of the oral microbiome, the Human Oral Microbiome Database (a resource for exploring the Oral Microbiome), and efforts to connect the oral metagenome with the oral metaproteomics and structural metagenomics."
Jumat, 06 Maret 2009
The ecosystem that is your stomach! Genomic and Genetic Insight into Gut Microbiota Function and Manipulation
"Trillions of microbes live in our digestive tract and influence our biology in profound and diverse ways. Several diseases, including obesity and inflammatory bowel diseases, have been associated with large-scale shifts in microbiota composition. The ability to address basic questions concerning community function and plasticity are fundamental to understanding the extent of causal relationships between host biology and microbiota perturbations, and whether the microbiota is a viable therapeutic target. One of our long-term goals is to achieve a level of functional understanding that, if provided the metagenome of an individual’s microbiota, would allow us to accurately predict how the microbial community will functionally adapt to a specific perturbation (e.g., dietary change). To investigate how changes in the intestinal environment alter microbiota function, and how these changes, in turn, influence host biology we have characterized responses of simplified microbiotas living within the gut of gnotobiotic mice to changes in host diet, community membership, and host genotype. These studies have revealed the importance of a finely-tuned system of polysaccharide sensing and utilization in the model symbiont Bacteroides thetaiotaomicron (B. theta). We are currently using a single polysaccharide utilization locus dedicated to dietary fructan utilization of B. theta as a model to understand mechanisms underlying diet-induced changes in microbiota function and composition. Genetic ablation of proteins involved in the multi-step process of sensing, harvesting, degrading, and metabolizing fructans variably cripples B. theta’s utilization of fructose-based polysaccharides depending upon which step of consumption is compromised. These findings are consistent with functional differences in fructan utilization between Bacteroides species. Together these results set the stage for predicting, based on gene content, how microbiotas respond to changes in the nutrient environment and suggest how metagenomics could facilitate personalized therapeutic manipulation of the microbiota."
A proposal for a field guide for microbes just like a field guide for birds. "A Genomic Encyclopedia of Bacteria and Archaea (GEBA)"
"There is a glaring gap in microbial genome sequence availability – the currently available genome sequences show a highly biased phylogenetic distribution compared to the extent of microbial diversity known today. This bias has resulted in major limitations in our knowledge of microbial genome complexity and our understanding of the evolution, physiology and metabolic capacity of microbes. Although there have been small efforts in sequencing genomes from across the tree of life for microbes, there are no systematic efforts. There are many reasons why phylogenetic based sequencing in theory should be of great benefit including: (a) improved identification of protein families and orthology groups across species, which will improve annotation of other microbial genomes (b) improved phylogenetic anchoring of metagenomic data, (c) gene discovery (which tends to be maximized by selecting phylogenetically novel organisms, (d) a better understanding of the processes underlying the evolutionary diversification of microbes (e.g., lateral gene transfer and gene duplication) (e) a better understanding of the classification and evolutionary history of microbial species and (f) improved correlations of phenotype and genotype in microbes. Based on the potential benefits, we (JGI) have commenced a pilot project to create a Genomic Encyclopedia of Bacteria and Archaea (GEBA). In this pilot, we plan to sequence ~100 genomes selected based on their phylogenetic novelty. This is being done at two phylogenetic scales. About 60 of the genomes are from across the breadth of bacteria and archaea. The remaining 40 genomes are from within the Actinobacteria. By doing this two tiered selection we can test both the value of breadth from across the bacteria and archaea as well as the value of filling in the phylogenetic gaps within a single phyla. In my talk I will summarize the project and report on the sequencing and analysis of the first 56 genomes. I will discuss how we are using this pilot to test protocols that could be used for a scale up of the GEBA project or for any other large scale microbial sequencing project. In addition I will discuss how collaborations with culture collections can be valuable in such a project. Finally, I will report on the results of tests of the value of phylogenetic based sequencing."
Marine Biologist
A little something for the weekend.
Combine this with UCSB Marine Biologist Milton Love's little essay on some good, and not so good, reasons to be a Marine Biologist.
Oh and can a golf ball really block the blowhole of a whale? You're CCS students, you should be able to take a crack at that question
A trip through some of the phylums of life: The biology of chordates, annelids, and cnidarians
Biology of annelids.
Biology of cnidarians
Kamis, 05 Maret 2009
Signs point to sponges
 Just a few weeks ago there was a nature paper based on 'chemical fossils': Fossil steroids record the appearance of Demospongiae during the Cryogenian period.
Just a few weeks ago there was a nature paper based on 'chemical fossils': Fossil steroids record the appearance of Demospongiae during the Cryogenian period.MIT, where some of the researchers are based, had a press release: Signs point to sponges as earliest animal life 'Chemical fossils' provide evidence for first multicelled creatures.
The Cryogenian is that period in the Precambrian era when there was an extensive series of severe ice ages - often referred to as 'snowball earth'.
Soft-bodied animals such as sponges are very rarely preserved as fossils, so finding evidence of their early appearance required some clever detective work. The key turned out to be an examination of unusual chemicals: steroids of a particular type produced abundantly by sponges but virtually never by simpler organisms.
Studying an unusually well preserved long sequence of strata found in Oman, the research team was able to extract these "chemical fossils" from a large number of samples spanning a range of tens of millions of years -- before, during and after the Ediacarian period. This provided clear evidence that sponges must have evolved long before the great variety of multicellular organisms that proliferated at the dawn of that period.
At that time in geological history, the Earth was just coming out of the last of its "snowball Earth" phases, when the entire planet was shrouded in ice. Since the new findings show that complex life seems to have begun tens of millions of years before that, that means these organisms were able to survive through that extreme episode of glaciation, something that many scientists had thought was impossible. This provides new evidence that the freezing was not absolute, but instead left some open patches of water.
"There's plenty of evidence in these rocks that there were places on Earth where life was flourishing" during this snowball episode, known as the Cryogenian, Summons says. "There must have been some refugia. Life certainly didn't shut down."
Rabu, 04 Maret 2009
Dave Issadore over at Harvard explains integrated circuts and shows you how he made cells do the waltz!
To learn more, check out their lab on a chip paper:
http://www.rsc.org/Publishing/Journals/LC/article.asp?DOI=b710928h
Dr. Jiang explains Lab-on-a-chip! Here he talks about controlled Microfluidic Interfaces for Microoptics and Microsensing (Video 2008 September)
EECS Department Colloquium (EECS 500) Hongrui Jiang, Ph.D.
"Controlled Microfluidic Interfaces for Microoptics and Microsensing"
September 11, 2008 Lab on a chip has found many applications in biological and chemical analysis. Because these labs on chips involve handling of fluids at the microscale, surface tension profoundly affects the behavior and performance of these systems.
Recombinant DNA
A 3D animation illustrating the process by which a protein is mass-produced using spliced DNA and bacterial replication.
Fossil Fish Brain
 The structure of the skull (foreground) of a 300-million-year-old iniopterygian fish from Kansas remotely related to living ratfish is elucidated thanks to holotomography, a technique based on synchrotron X-ray phase contrast imaging (background), and yields the first hint at an exceptional mineralization of the brain (orange). (Credit: PNAS/Philippe Janvier (CNRS, Museum National d'Histoire Naturelle))
The structure of the skull (foreground) of a 300-million-year-old iniopterygian fish from Kansas remotely related to living ratfish is elucidated thanks to holotomography, a technique based on synchrotron X-ray phase contrast imaging (background), and yields the first hint at an exceptional mineralization of the brain (orange). (Credit: PNAS/Philippe Janvier (CNRS, Museum National d'Histoire Naturelle))I think this must be the 300 million year old fish brain that was mentioned on Tuesday. Although it is the first time that the soft tissue of such an old fossil brain has ever been found it sounds like it is also one of the first times people have used sophisticated imaging techniques to look inside the fossil skull.
(S)cientists used the technique of absorption microtomography to study different samples. One sample, stemming from Kansas (US), revealed a peculiar structure: it was denser than the surrounding matrix that fills the braincase, and which is made of crystalline calcite. In order to elucidate its structure in detail, they decided to use a second technique, X-ray holotomography. Surprisingly, the results showed a symmetrical and elongated object placed in the same position as a brain would have been.
Designer Babies? - Not Yet!
The unresolved issue of whether would-be parents should be allowed to create designer babies is a train wreck waiting to happen. New York, for example, does not have laws against using PGD strictly for cosmetic purposes, so clinics such as Dr. Steinberg’s are free to promise what they will. Nevertheless, it’s a slippery ethical slope they’re on. The issue is discussed in Human Biology 5th ed., pp. 396-397.
Intro to the biology of plants (Video 1997) - Warning 90's era video
Selasa, 03 Maret 2009
Carbon dating and global warming
 The prodigious mobilization of science that produced nuclear weapons was so far-reaching that it revolutionized even the study of ancient climates. Nuclear laboratories, awash with funds and prestige, spun off the discovery of an amazing new technique — radiocarbon dating.
The prodigious mobilization of science that produced nuclear weapons was so far-reaching that it revolutionized even the study of ancient climates. Nuclear laboratories, awash with funds and prestige, spun off the discovery of an amazing new technique — radiocarbon dating. The discovery of carbon dating is an interesting story in its own right and is described in several places, for example here. It is also described here, at the American Institute of Physics, part of a rather nice collection of essays on the discovery of global warming. The ratio of C-14 to C-12 on earth has varied significantly during the Earth's history. This variation is due to changes in the intensity of the cosmic radiation bombardment of the Earth, and changes in the effectiveness of the atmosphere in deflecting that bombardment. To compensate for this variation, dates obtained from radiocarbon laboratories are now corrected using standard calibration tables.
It was particularly interesting that, as Stuiver had suspected, the carbon-14 wiggles correlated with long-term changes in the number of sunspots. Turning it around, Suess remarked that "the variations open up a fascinating opportunity to perceive changes in the solar activity during the past several thousand years." The anomalies were evidence for something that many scientists found difficult to believe — the surface activity of the Sun had varied substantially in past millennia. Carbon-14 might not only provide dates for long-term climate changes, but point to one of their causes.
Researchers make stem cell breakthrough - STEM CELLS WITHOUT EMBRYOS!
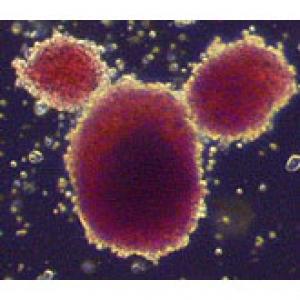
QUOTED FROM: PSYSORG.com http://www.physorg.com/news155138024.html
"This new method of generating stem cells does not require embryos as starting points and could be used to generate cells from many adult tissues such as a patient's own skin cells." - Dr. Nagy, Senior Investigator at the Samuel Lunenfeld Research Institute of Mount Sinai Hospital, Investigator at the McEwen Centre for Regenerative Medicine, and Canada Research Chair in Stem Cells and Regeneration.
Dr. Nagy discovered a new method to create pluripotent stem cells (cells that can develop into most other cell types) without disrupting healthy genes. Dr. Nagy's method uses a novel wrapping procedure to deliver specific genes to reprogram cells into stem cells. Previous approaches required the use of viruses to deliver the required genes, a method that carries the risk of damaging the DNA. Dr. Nagy's method does not require viruses, and so overcomes a major hurdle for the future of safe, personalized stem cell therapies in humans.
"This research is a huge step forward on the path to new stem cell-based therapies and indicates that researchers at the Lunenfeld are at the leading edge of regenerative medicine," said Dr. Jim Woodgett, Director of Research for the Samuel Lunenfeld Research Institute of Mount Sinai Hospital. Regenerative medicine refers to enabling the human body to repair, replace, restore and regenerate its own damaged or diseased cells, tissues and organs.
The research was funded by the Canadian Stem Cell Network and the Juvenile Diabetes Research Foundation (United States).
Dr. Nagy joined Mount Sinai Hospital as a Principal investigator in 1994. In 2005, he created Canada's first embryonic stem cell lines from donated embryos no longer required for reproduction by couples undergoing fertility treatment. That research played a pivotal role in Dr. Nagy's current discovery.
One of the critical components reported in Nagy's paper was developed in the laboratory of Dr. Keisuke Kaji from the Medical Research Council (MRC) Centre for Regenerative Medicine at the University of Edinburgh. Dr. Kaji's findings are also published in the March 1, 2009 issue of Nature. The two papers are highly complementary and further extend Nagy's findings.
"I was very excited when I found stem cell-like cells in my culture dishes. Nobody, including me, thought it was really possible," said Dr. Kaji. "It is a step towards the practical use of reprogrammed cells in medicine."
Source: Samuel Lunenfeld Research InstituteUnderstanding the classification of life: What is a Phylum?

WIKI: A phylum (plural: phyla)[note 1] is a taxonomic rank below Kingdom and above Class. "Phylum" is equivalent to the botanical term division.[1]
Although a phylum is often spoken of as if it were a hard and fast entity, no satisfactory definition of a phylum exists. Consequently the number of phyla varies from author to author. The relationship of phyla is increasingly well known, and larger clades can be erected to contain many of the phyla.
Informally, phyla can be thought of as grouping animals based on general body plan,[2] developmental or internal organizations.[3] For example, though seemingly divergent, spiders and crabs both belong to Arthropoda, whereas earthworms and tapeworms, similar in shape, are from Annelida and Platyhelminthes, respectively. Although the International Code of Botanical Nomenclature allows the use of the term "phylum" in reference to plants, the term "Division" is almost always used by botanists.
The best known animal phyla are the Mollusca, Porifera, Cnidaria, Platyhelminthes, Nematoda, Annelida, Arthropoda, Echinodermata, and Chordata, the phylum to which humans belong. Although there are approximately 35 phyla, these nine include over 96% of animal species. Many phyla are exclusively marine, and only one phylum, the Onychophora (velvet worms) is entirely absent from the world's oceans–although ancestral oncyophorans were marine.[4]
MARINE BIO: INTRO TO Echinoderms (Phylum: Echinodermata)

WIKI: Echinoderm
Fossil range: Cambrian - present
Domain: Eukaryota
Kingdom: Animalia
Subkingdom: Eumetazoa
Superphylum: Deuterostomia
Phylum: Echinodermata
Echinoderms (Phylum Echinodermata) are a phylum of marine animals (including sea stars). Echinoderms are found at every ocean depth, from the intertidal zone to the abyssal zone. Aside from the problematic Arkarua, the first definitive members of the phylum appeared near the start of the Cambrian period. The phylum contains about 7,000 living species, making it the second-largest grouping of deuterostomes, after the chordates; they are also the largest phylum that has no freshwater or terrestrial representatives. The word derives from the Greek εχινοδέρματα (echinodermata), plural of εχινόδερμα (echinoderma), "spiny skin" and that from εχινός (echinos), "sea-urchin", originally "hedgehog"[1] + δέρμα (derma), "skin"[2][3]. The Echinoderms are important both biologically and geologically: biologically because few other groupings are so abundant in the biotic desert of the deep sea, as well as the shallower oceans, and geologically as their ossified skeletons are major contributors to many limestone formations, and can provide valuable clues as to the geological environment. Further, it is held by some that the radiation of echinoderms was responsible for the Mesozoic revolution of marine life. Two main subdivisions of Echinoderms are traditionally recognised: the more familiar, motile Eleutherozoa, which encompasses the Asteroidea (starfish), Ophiuroidea (brittle stars), Echinoidea (sea urchins and sand dollars) and Holothuroidea (sea cucumbers); and the sessile Pelmatazoa, which consists of the crinoids. Some crinoids, the feather stars, have secondarily re-evolved a free-living lifestyle. A fifth class of Eleutherozoa consisting of just two species, the Concentricycloidea (sea daisies), were recently[4] merged into the Asteroidea. The fossil record contains a host of other classes which do not appear to fall into any extant crown group.

